Abstract
FLT3 internal tandem duplication mutations (FLT3-ITD) drive both pediatric and adult AML, but it cooperates with different mutations at different ages. For example, interactions between FLT3-ITD and NUP98 translocations are more common in childhood AML, whereas interactions between FLT3-ITD and RUNX1, DNMT3A or IDH1/2 mutations are more common in adult AML. These differences raise the question of whether FLT3-ITD drives distinct transcriptional programs when paired with different, age-biased cooperating mutations, and whether such distinctions convey unique, cooperating mutation-specific vulnerabilities.
To answer these questions, we first compared the transcriptional and epigenetic changes that define FLT3-ITD/Nup98-HoxD13 (NHD13) and FLT3-ITD/Runx1-deleted (FLT3-ITD/Runx1del) hematopoietic progenitors. The FLT3-ITD/NHD13 model reflects a pediatric-like mutation profile, whereas the FLT3-ITD/Runx1del model reflects an adult-like mutation profile. Both mutation profiles markedly altered hematopoiesis and HSC/MPP function. However, RNA-seq data and Chipmentation revealed non-overlapping cooperative changes in gene expression and enhancer activation between the two genotypes. FLT3-ITD/NHD13 mutations selectively induced type I interferon (IFN-1) target gene expression in a cooperative manner. In contrast, FLT3-ITD/ Runx1del selectively induced Mir155, a negative regulator of IFN-1. Thus, the mutations engage distinct IFN-1 responses.
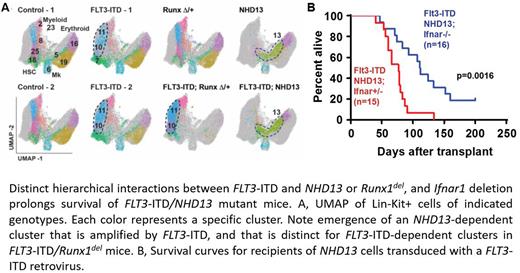
To further understand how the mutations reprogram hematopoiesis, we performed Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-seq) on Lin-Kit+ progenitors. FLT3-ITD by itself induced expansion of myeloid-biased multipotent progenitor populations (MPPs), and FLT3-ITD/ Runx1del cooperation markedly amplified the changes. In contrast, NHD13 reprogrammed MPPs, and FLT3-ITD/NHD13 amplified this emergent population (Figure A). These results reveal distinct hierarchical interactions between FLT3-ITD and its cooperating mutations, with FLT3-ITD/ Runx1del progenitors closely resembling FLT3-ITD progenitors (i.e., the FLT3-ITD was dominant), and FLT3-ITD/NHD13 progenitors closely resembling NHD13 progenitors (i.e., the NHD13 was dominant). These interactions appeared to affect sensitivity to the FLT3-ITD inhibitor gilteritinib, as FLT3-ITD/ Runx1del progenitors were suppressed by gilteritinib while FLT3-ITD/NHD13 progenitors were not.
We evaluated the role of IFN-1 signaling in sustaining FLT3-ITD/NHD13 progenitors and in promoting AML using additional CITE-seq, transplantation assays and survival studies. Deleting the IFN-1 receptor (Ifnar1) suppressed a specific subpopulation of CD317+Lin-Kit+ progenitors in FLT3-ITD/NHD13 mice, and it reduced expression of several genes that can drive leukemogenesis, including Cdk6, Gata2 and Msi2. Ifnar1 deletion significantly prolonged survival of mice of FLT3-ITD/NHD13 mice (Figure B).
Altogether, these data show that FLT3-ITD can drive distinct transcriptional programs when paired with different cooperating mutations. Furthermore, differences between pediatric and adult mutation profiles can potentially convey distinct sensitivity to FLT3-ITD inhibition while unmasking age-specific gene dependencies and therapeutic vulnerabilities. IFN-1 is one such dependency in NUP98-translocated AML initiation.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal